COVID infections and the link to the gut microbiome
Recent research into COVID-19 has found that the composition of the gut microbiome may influence the severity and duration of infections and how the immune system responds.
COVID-19 is primarily a respiratory illness, but gastrointestinal symptoms, including nausea, abdominal pain, vomiting and diarrhoea are also commonly reported: indicating the involvement of the gut.
The COVID-19 virus invades cells by a process that starts with the virus binding to a cell surface receptor called ACE2 (Gao et al., 2020). While this receptor is found in high levels on the surfaces of cells lining the lungs (lung epithelium), it is also found in high levels in the intestines (intestinal epithelium) (Uno, 2020).
In the gut, ACE2 is linked to the gut microbiome and plays a role in gut inflammation (Gao et al., 2020; Garg et al., 2020).
COVID-19 viral material has been detected in stool samples, even after respiratory tract samples test negative. This all shows that the COVID-19 virus can and does invade the intestinal epithelium.
In order for the COVID-19 virus to invade intestinal epithelial cells, it must survive transit through the stomach, and resist stomach acid. Individuals on proton pump inhibitors (which reduce stomach acidity) are at risk of increased COVID-19 symptom severity and duration (Lee et al., 2021; Lee et al., 2020; Zhou et al., 2020).
Similarly, susceptibility to the virus is age-related (Zimmerman & Curtis, 2020), and increased age is associated with decreased stomach acidity (Uno, 2020), as well as decreased overall immune function and microbial dysbiosis (Amsterdam & Ostrov, 2018).
The makeup of the gut microbiome also plays a role in COVID-19 incidence and severity.
In a recent study, researchers from the Chinese University of Hong Kong (Yeoh et al., 2021) analysed blood and stool samples and medical records from 100 people with COVID-19 infections, and 78 people without COVID-19 who were taking part in a microbiome study before the pandemic.
Analysis of the stool samples showed that the make-up of the gut microbiome differed significantly between patients with and without COVID-19, irrespective of whether they had been treated with drugs, including antibiotics.
COVID-19 patients had higher numbers than people without the infection of several bacterial species including Ruminococcus gnavus, Ruminococcus torques and Bacteroides dorei. The COVID-19 patients also had far fewer of the bacterial species that have been shown to improve immune system responses, such as Bifidobacterium adolescentis, Faecalibacterium prausnitzii (F. prau) and Eubacterium rectale. Lower numbers of F. prau and Bifidobacterium bifidum were particularly associated with the severity of COVID-19 symptoms.
A previous study showed COVID-19 patients had lower numbers of several bacterial species, including E. rectale and F. prau compared to healthy individuals (Zuo et al., 2020).
Again, F. prau, and a bacterium called Alistipes, most strongly negatively correlated with disease severity. These COVID-19 patients were also shown to have greater numbers of opportunistic pathogens known to cause bacteraemia (presence of bacteria in the bloodstream), and greater numbers of species known to upregulate ACE2 numbers in the gut. This microbial dysbiosis persisted during and after hospitalisation of the patients. A further study investigated gut microbiome differences in patients with COVID-19 and influenza A (H1N1) compared to healthy people (Gu et al., 2020).
People with COVID-19 or H1N1 had lower microbial diversity (a measure of microbial ecosystem robustness), and the relative abundance of Streptococcus and Escherichia/Shigella was significantly higher in COVID-19 and H1N1 patients, respectively. A lower relative abundance of beneficial gut microbes, including Faecalibacterium was also observed in both COVID-19 and H1N1 patients.
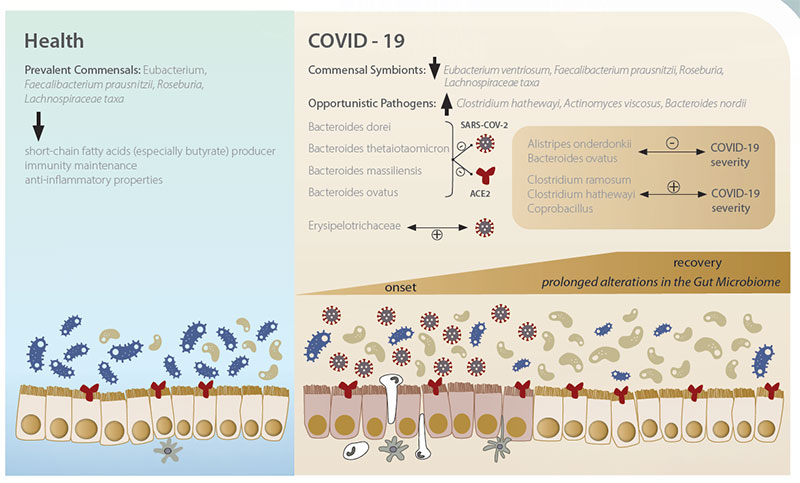
Livaux and F. prau severity fact sheet – from Zuo et al. Alterations in gut microbiota of patients with Covid-19 during time of hospitalisation. Gastroenterology. 2020. 159: 944-955
The depletion of key bacterial species in the gut microbiota of COVID-19 patients was also associated with increased concentrations of inflammatory cytokines (Yeoh et al., 2021), suggesting the gut microbiome is influencing the immune system’s response to the COVID-19 infection. Imbalances in the make-up of the microbiome may also be implicated in persistent inflammatory symptoms, dubbed ‘long COVID’.
The collective research suggests that bolstering of beneficial gut species depleted in COVID-19 such as F. prau through the use of prebiotics and/or probiotics could serve as a way to reduce the duration and severity of the disease.
However, F. prau is highly oxygen sensitive and cannot be viably delivered for consumption in adequate live numbers in a probiotic supplement format.
A better proposition are prebiotics: food which survives digestion and reaches our large intestine to selectively increase the numbers of gut bacteria that confer health benefits. Diet has been shown to increase F. prau numbers (Singh et al., 2017), particularly high carbohydrate/low glycemic impact diets (Fava et al., 2013).
In terms of selectively increasing F. prau, Livaux® from New Zealand gold kiwifruit is a natural prebiotic clinically shown to increase F. prau levels in individuals with low baseline levels (Blatchford et al., 2017).
This effect has also been demonstrated in vitro (Duysburgh et al., 2019). Livaux contains high methoxy pectin, which is known to be a substrate (food) used for growth by F. prau (Lopez-Siles et al., 2011).
References
Amsterdam, D., & Ostrov, B. E. (2018). The impact of the microbiome on immunosenescence. Immunological Investigations, 47(8), 801-811.
Blatchford, P., Stoklosinski, H., Eady, S., Wallace, A., Butts, C., Gearry, R., … & Ansell, J. (2017). Consumption of kiwifruit capsules increases Faecalibacterium prausnitzii abundance in functionally constipated individuals: a randomised controlled human trial. Journal of Nutritional Science, 6.
Duysburgh, C., Van den Abbeele, P., Krishnan, K., Bayne, T. F., & Marzorati, M. (2019). A synbiotic concept containing spore-forming Bacillus strains and a prebiotic fiber blend consistently enhanced metabolic activity by modulation of the gut microbiome in vitro. International journal of pharmaceutics: X, 1, 100021.
Fava, F. R. A. N. C. E. S. C. A., Gitau, R., Griffin, B. A., Gibson, G. R., Tuohy, K. M., & Lovegrove, J. A. (2013). The type and quantity of dietary fat and carbohydrate alter faecal microbiome and short-chain fatty acid excretion in a metabolic syndrome ‘at-risk’population. International journal of obesity, 37(2), 216-223.
Gao, Q. Y., Chen, Y. X., & Fang, J. Y. (2020). 2019 Novel coronavirus infection and gastrointestinal tract. Journal of digestive diseases, 21(3), 125.
Garg, M., Christensen, B., & Lubel, J. S. (2020). Gastrointestinal ACE2, COVID-19 and IBD: Opportunity in the Face of Tragedy?. Gastroenterology, 159(4), 1623-1624.
Gu, S., Chen, Y., Wu, Z., Chen, Y., Gao, H., Lv, L., … & Li, L. (2020). Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1N1 influenza. Clinical Infectious Diseases, 71(10), 2669-2678.
Lee, S. W., Ha, E. K., Yeniova, A. Ö., Moon, S. Y., Kim, S. Y., Koh, H. Y., … & Yon, D. K. (2021). Severe clinical outcomes of COVID-19 associated with proton pump inhibitors: a nationwide cohort study with propensity score matching. Gut, 70(1), 76-84.
Lee, S. W., Yang, J. M., Yoo, I. K., Moon, S. Y., Ha, E. K., Yeniova, A. Ö., … & Yon, D. K. (2020). Proton pump inhibitors and the risk of severe COVID-19: a post-hoc analysis from the Korean nationwide cohort. Gut.
Lopez-Siles, M., Khan, T. M., Duncan, S. H., Harmsen, H. J., Garcia-Gil, L. J., & Flint, H. J. (2012). Cultured representatives of two major phylogroups of human colonic Faecalibacterium prausnitzii can utilize pectin, uronic acids, and host-derived substrates for growth. Applied and environmental microbiology, 78(2), 420-428.
Singh, R. K., Chang, H. W., Yan, D. I., Lee, K. M., Ucmak, D., Wong, K., … & Liao, W. (2017). Influence of diet on the gut microbiome and implications for human health. Journal of translational medicine, 15(1), 1-17.
Zuo, T., Zhang, F., Lui, G. C., Yeoh, Y. K., Li, A. Y., Zhan, H., … & Ng, S. C. (2020). Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology, 159(3), 944-955.
Uno, Y. (2020). Why does SARS-CoV-2 invade the gastrointestinal epithelium? Gastroenterology, 159(4), 1622-1623.
Yeoh, Y. K., Zuo, T., Lui, G. C. Y., Zhang, F., Liu, Q., Li, A. Y., … & Ng, S. C. (2021). Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut, 70(4), 698-706.
Zhou, J., Wang, X., Lee, S., Wu, W. K. K., Cheung, B. M. Y., Zhang, Q., & Tse, G. (2020). Proton pump inhibitor or famotidine use and severe COVID-19 disease: a propensity score-matched territory-wide study. Gut.
Zimmermann, P., & Curtis, N. (2021). Why is COVID-19 less severe in children? A review of the proposed mechanisms underlying the age-related difference in severity of SARS-CoV-2 infections. Archives of disease in childhood, 106(5), 429-439.